WP1 – Ecoinformatics
Research Team
- Prof. Alex J Dumbrell (Work Package Lead) - University of Essex
- Dr Boyd McKew - University of Essex
- Dr Robert Ferguson - University of Essex
- Dr Kate Randall - University of Essex
Aims: a) define the chemicals that are on the horizon as emerging threats; b) to identify experimental modules and field sites where our approach can be tested and validated; c) support other work packages by producing NGS data describing how taxonomic and functional biodiversity responds to chemical stressors.
Methods: Identifying the key drivers and effective response variables through mining existing chemical and biotic data (e.g. Defra, EA, RP and other databases). We will start by exploring existing data to characterise the current and emerging “exposome” of chemicals. We will create a relational database comprising possible chemical stressors and their reported biological modes of action. Initially, chemicals close-to-market will be prioritised, starting with pharmaceutical and pesticide stressors identified in the EU Water Framework Directive and UK Red List. Data collection will target large EA, RP and EU databases, NERC repositories, other open-access sources (e.g. Dryad) and text-mining peer reviewed papers, policy reports and websites. Using a “Big Data” approach, we will group chemicals based on biological modes of action and related observed ecological responses. This will allow us to reduce the multidimensionality (i.e. the breadth of unique chemicals with similar ecological impacts) and select representatives for our experimental phase (see WP2, WP3 & WP4).
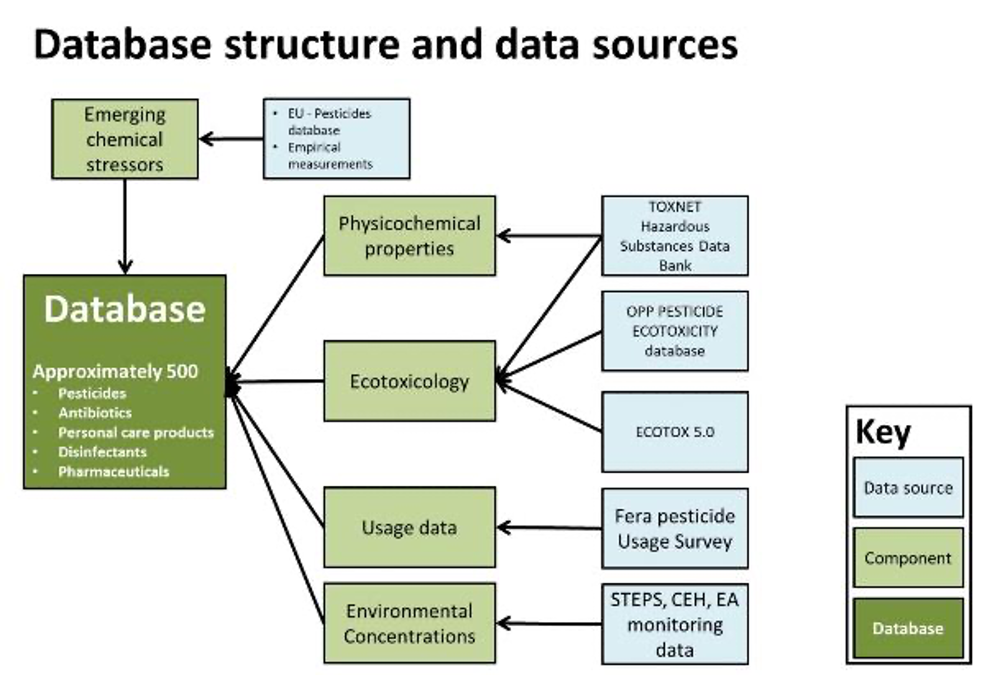 Emerging chemicals database structure and data sources. Emerging chemical stressors are identified from existing databases and monitoring data. Then a number of data sources are used to determine the environmental risk of the chemicals, such as ecotoxiclogical data for specific taxa, which is compared with predicted environmental concentrations based on monitoring data and modelling.
Emerging chemicals database structure and data sources. Emerging chemical stressors are identified from existing databases and monitoring data. Then a number of data sources are used to determine the environmental risk of the chemicals, such as ecotoxiclogical data for specific taxa, which is compared with predicted environmental concentrations based on monitoring data and modelling.
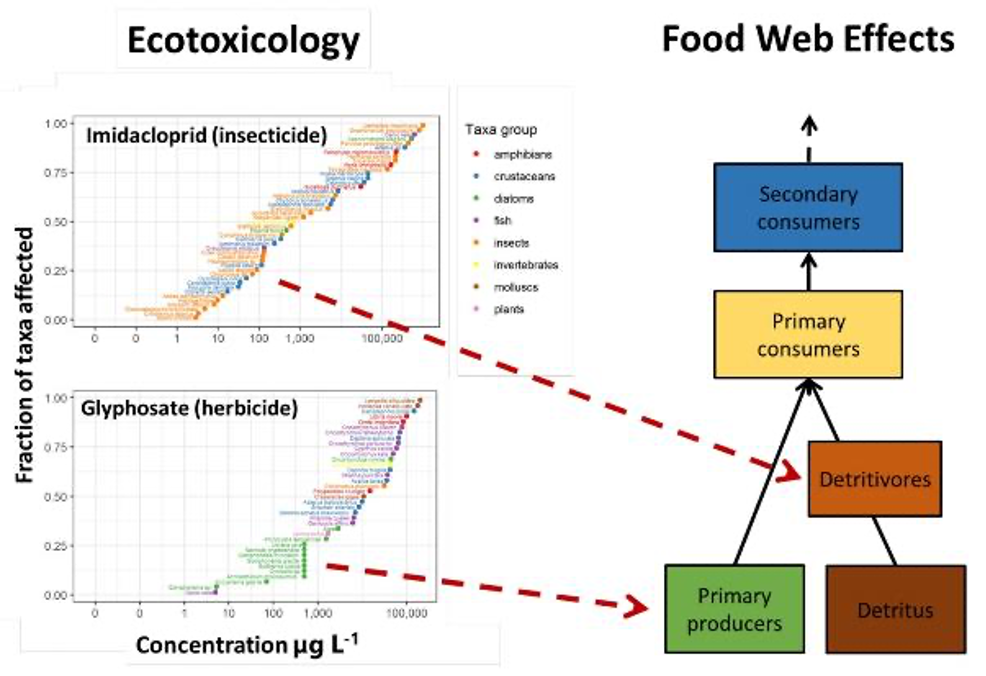 Predicting food web effects from ecotoxicology data. Species sensitivity distributions are calculated from toxicity data for multiple taxa. This can then be used to predict the effects of individual chemicals in the food web and predict cascading indirect effects.
Predicting food web effects from ecotoxicology data. Species sensitivity distributions are calculated from toxicity data for multiple taxa. This can then be used to predict the effects of individual chemicals in the food web and predict cascading indirect effects.
The other core task for WP1 is producing molecular (NGS/qPCR) data resulting from samples collected in experiments and field campaigns (WP2-4). Here we generate the taxonomic and functional biodiversity measures describing community dynamics, resilience and issues of functional redundancy in the microbial (and macro) components of the food web. We use NGS metabarcoding to quantify taxonomic/phylogenetic diversity and NGS metagenomics and qPCR to quantify microbial functional responses. Full details of these molecular methods can be found here