WP4 – Ecological networks and field bioassays
Research Team
- Prof. Guy Woodward (Work Package Lead) - Imperial College London
- Dr Eoin O’Gorman - University of Essex
- Dr Michelle Jackson - University Oxford
- Dr Danielle Harris - Imperial College London
Aim
Extend our lab and mesocosm experiments (WP3 into natural settings to test for consistency of responses
Method
We cannot dose natural systems with chemicals so we will collect samples from rivers across the UK (Figure 1), spanning a gradient of urban-pastoral-arable pollution. We will focus on sites where there is strong evidence of biocide effects beyond those associated with organic pollution (e.g. from wastewater). At our field sites, we will measure:
- Ecosystem processes (e.g. biomass production and decomposition rates) by deploying the same bioassays as in WP3 (cellulose and leaf litter degradation, algal tile colonisation)
- Abundance and biomass per unit area of all levels of the food web, from microbes to fishes
- Ecosystem-level variables (e.g. the efficiency of energy transfer through food webs) Examples of data generated by bioassays and field surveys are shown in Figure 2.
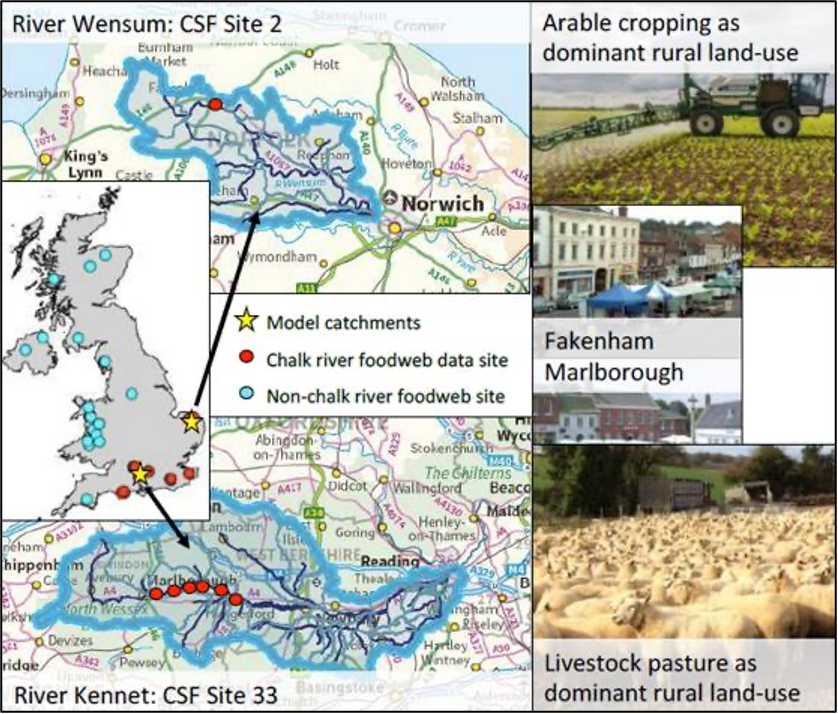
Fig. 1 Location of sites where we have detailed food web data across the UK, and the candidate model catchments: the Wensum (top) and the Kennet (bottom). Chalk rivers are the most biodiverse and productive of UK running waters, yet are also exposed to a range of chemical stressors in both rural and urban settings. The Kennet is predominantly pasture, whereas the Wensum is arable and monitored intensively for pesticides. Both are DEFRA/EA Catchment Sensitive Farming Priority Catchments. The model systems are of similar size, geology, urban-rural balance and spatial configurations. We already have highly resolved food web data, traditional biomonitoring data and chemical profiles for sites in each catchment
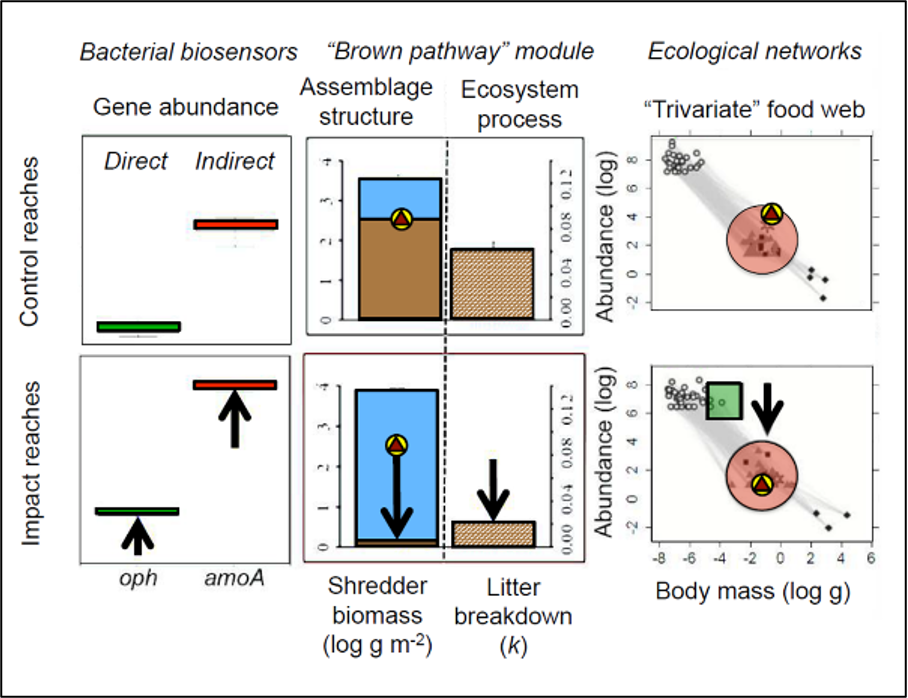
Fig. 2 Proof of concept of our gene-to-ecosystem framework. The impacts of a pesticide spill in the River Kennet, affecting multiple levels of the freshwater food web. The left-hand panel shows that oph and amoA gene abundances were increased after the spill. These genes are responsible for associated with the catalysis of organophosphate and the anaerobic breakdown of invertebrate carcases, respectively. The mid-panels indicate that leaf litter breakdown was reduced after the spill, due to a large reduction in the biomass of Gammarus pulex (river shrimp; brown triangle in yellow circle)